Request and download rich multi-omic data for Champions Oncology’s PDX bank and cell lines
Data Licensing allows Lumin users to use Tokens to request multi-omic data, which can be downloaded into their own environment for further analysis.
After opening Data Licensing from the Lumin Homepage user’s simply complete the report for to request the data for their models of interest. 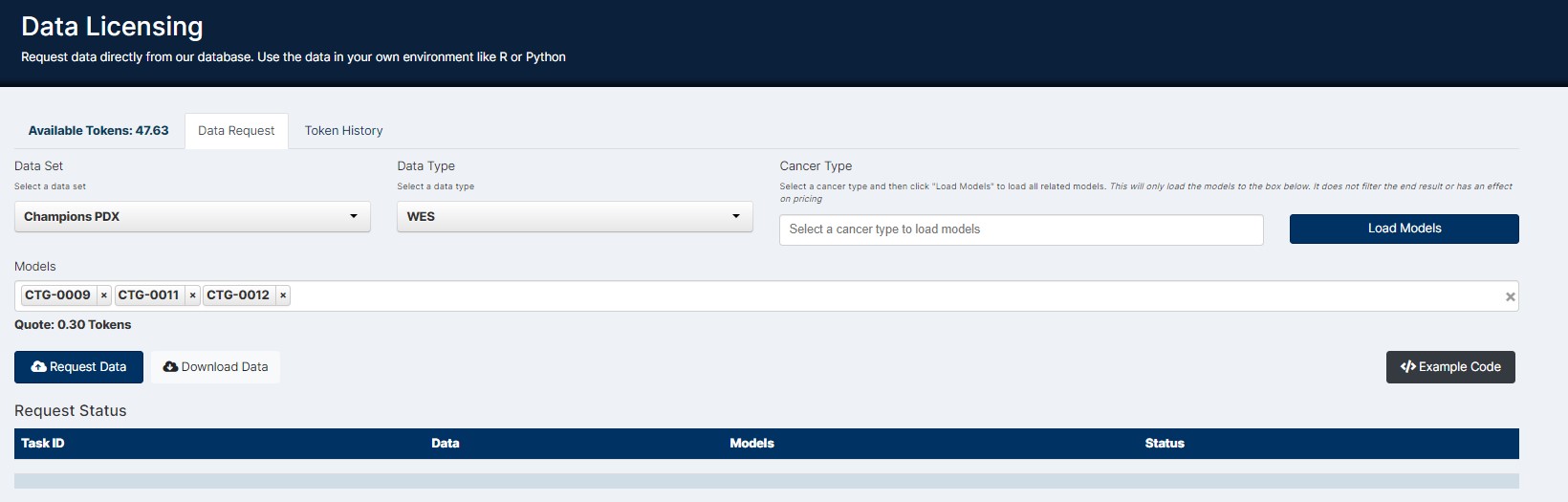
| File Name | Topics |
|---|---|
| ccle_heatmap__R.ipynb | Importing data from the Broad Institute’s depmap portal; hierarchical clustering |
| correlation_between_genes_R.ipynb | Correlation between genes regarding the expression data |
| dge_cancer_types_R.ipynb | Differential gene expression between cancer types |
| differential_protein_expression_R.ipynb | Normalization of proteomic data; volcano plot of fold change and P values |
| gene_signature_R.ipynb | Gene signature regarding the treated and naive groups |
| gsea_R.ipynb | Pathway enrichment analysis |
| gsva_kegg_pathways_in_R.ipynb | Importing pathways from MSigDB; GSVA analysis; DGE with limma |
| pca_ica_umap_R.ipynb | Comparing PCA, ICA, and UMAP for the gene-expression data |
| sensitivity_profiles_Achilles_Data_R.ipynb | Importing data from the Broad Institute’s depmap portal; static plotting of sensitivity scores |
| ssGSEA_R.ipynb | Single-sample pathway enrichment analysis |
| t-SNE_R.ipynb | Heatmap and t-SNE of the gene-expression data |
| wgcna_R.ipynb | Gene co-expression analysis |
| interative_heatmap_Python.ipynb | Enabling interactive widgets; creating clustered heatmaps |