Find differentially expressed genes across model cohorts.
Our Target & Biomarker Discovery tool aims to help users perform gene expression analysis, quantify mutational occurrences, pathway analysis or perform spearman/pearson correlations using Champions PDX or datasets run external to Champions. You can leverage existing in vivo %TGI for SOC agents to develop biomarker hypotheses. Users can upload their pharmacological drug study data (IC50 or TGI values) or subset models into group A versus group B. Users can compare datasets to identify differentially expressed genes or genes positively or negatively correlated with response.
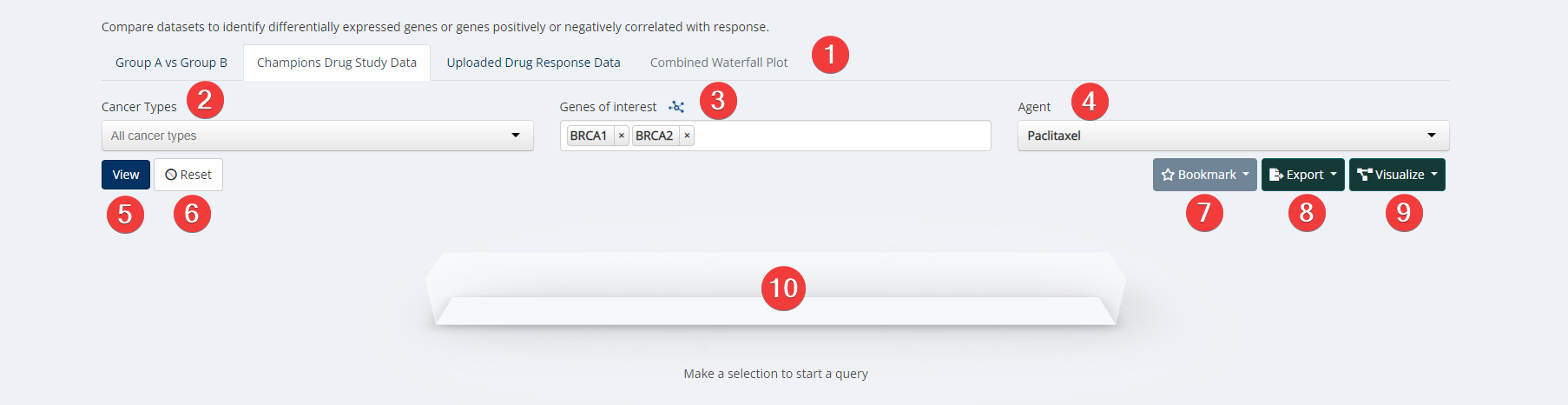
Target & Biomarker Discovery Input
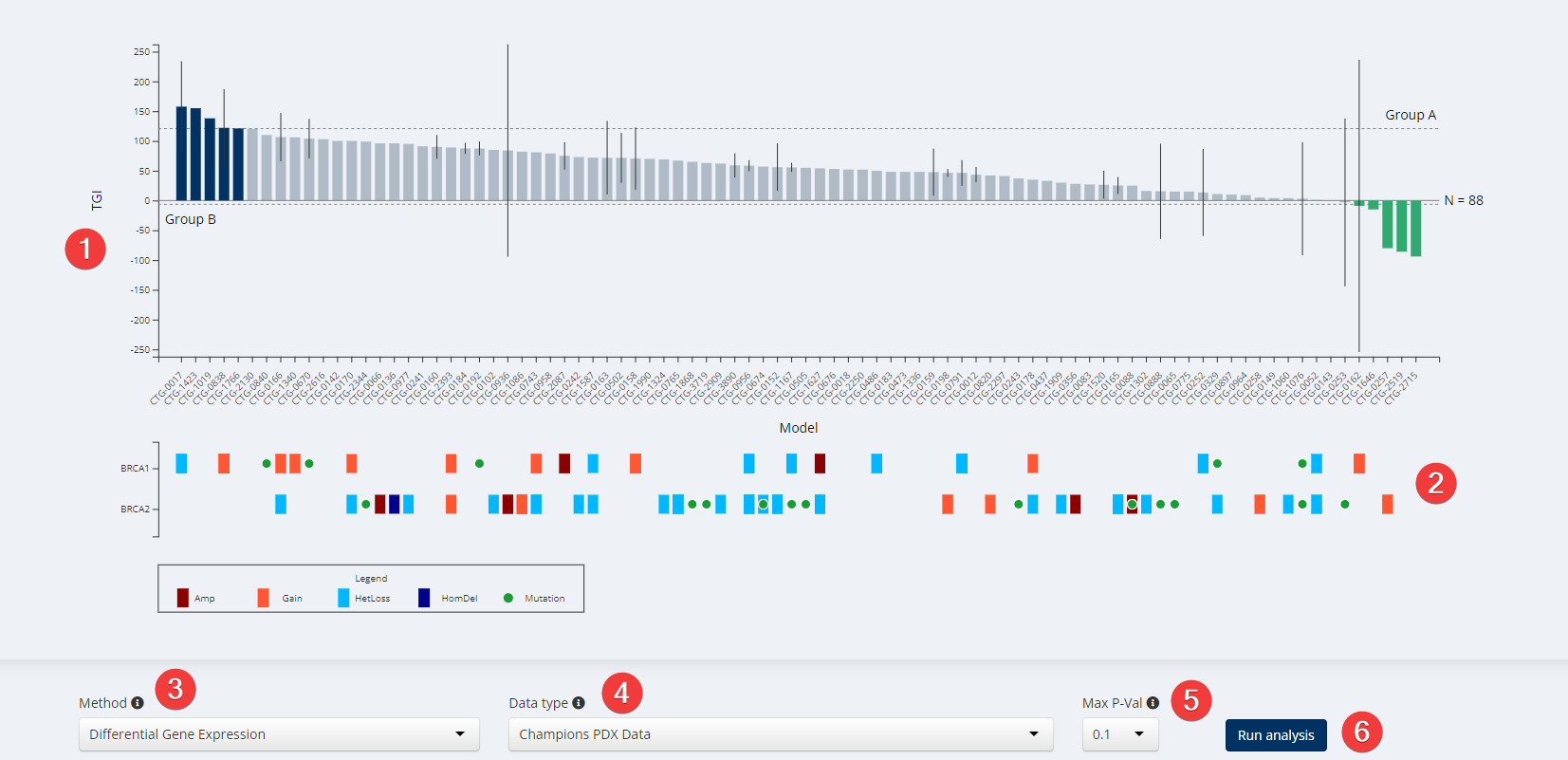
Target & Biomarker Discovery Visualization
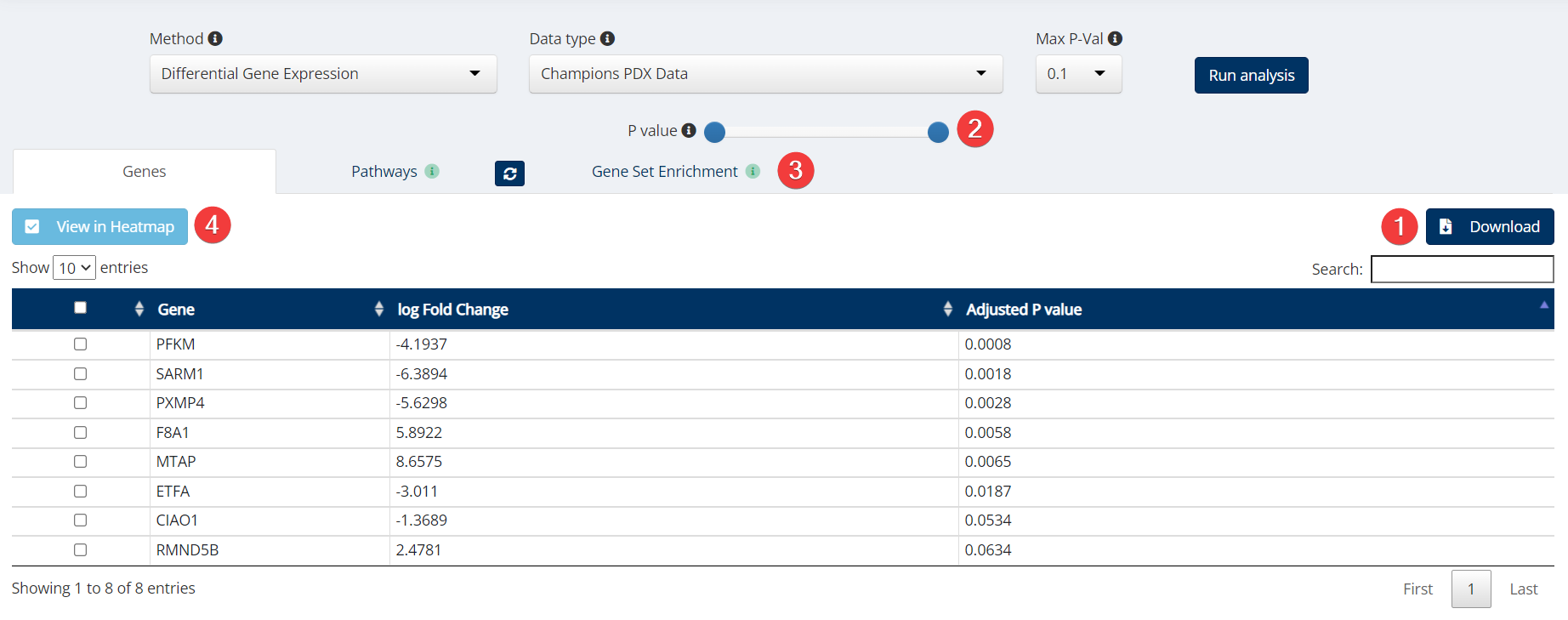
Differential Gene Expression
Differential Gene Expression (DGE) Analysis is done using the computation and ranking algorithms in edgeR with the default Benjamini & Hochberg (1995) p-value adjustment to correct the false discovery rate for multiple comparisons. The full analysis consists of fitting a linear model for each gene to the selected models in each group and then computing moderated t-statistics, moderated F-statistic, and log-odds of differential expression by empirical Bayes moderation of the standard errors towards a global value. The moderated t-statistics test each individual contrast is equal to zero, the moderated F-statistic tests whether all the contrasts are zero, and the log-odds of differential expression (B-statistic) can be used to infer whether the gene is differentially expressed. The log2-fold-change estimate is also reported for each contrast.
Spearman’s and Pearson’s correlation analyses compute the correlation between gene expression and TGI for a selected model group. Select a model group using one or both of the horizontal dashed lines on the waterfall plot. Unlike the DGE and Fisher’s analyses, the correlation analyses combine Groups A and B into a single group for analysis. Alternatively you may slide one threshold line beyond the bars and only use the other one to highlight a selected group. Which line you use depends on whether you are looking for correlations with the high response group (Group A when viewing TGI) or the low response group (Group B when viewing TGI in the waterfall).
Pearson’s product-moment correlation computes a correlation coefficient (cor) between the TGI and expression, while Spearman’s Rank-Order correlation computes the rho value. The Pearson method assumes the data are ratio or interval data and that the errors are normally distributed around the trendline, while the Spearman method assumes the data are ordinal and the errors are nonparametric. Both methods assume a monotonic relationship in the data.
The Fisher’s Exact test in Lumin creates a 2x2 contingency table for each gene+sample combination, filling the table with mutation count and wild-type count for each group. The test uses the null hypothesis that mutations for the gene are evenly distributed among samples. The output contains the gene symbol, a p-value indicating whether the null hypothesis can be rejected for that gene (and by extension whether we can be confident that the proportion of mutations in the two groups is different for some non-random reason), and the mutation counts in group A and group B. This test is useful for evaluating whether the occurance of mutations in both small and large sample groups is random or the product of a shared factor. In Lumin this test is performed in R using the stats::fisher.test() function.
| Alteration | Copy Number |
|---|---|
| Deep Deletion (HomDel) | 0 |
| Shallow Deletion (HetLoss) | 1 |
| Low Level Gain (Gain) | 3 |
| High Level Amplification (Amp) | >= 4 |
Champions PDX Data
Your Data
Develop a New Biomarker Hypothesis
Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B, 57, 289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x. https://www.jstor.org/stable/2346101.