Calculate, analyze, and visualize multi-well plate drug combination dose response and synergy.
Our Pharmacology Analysis tool aims to allow users to upload plate maps from drug response studies to compute IC50 values and create dose-response curves, and also analyze your combination studies by uploading data to simultaneously perform Loewe, Bliss, ZIP or HSA measurements of synergy, additivity or antagonism. Synergy results are presented as a table, heat map and 3D surface plot.
You can collaborate with colleagues by sharing your data files. Please Note: This is not an automatic feature, you need to check a box first to make the analysis visible to your company.
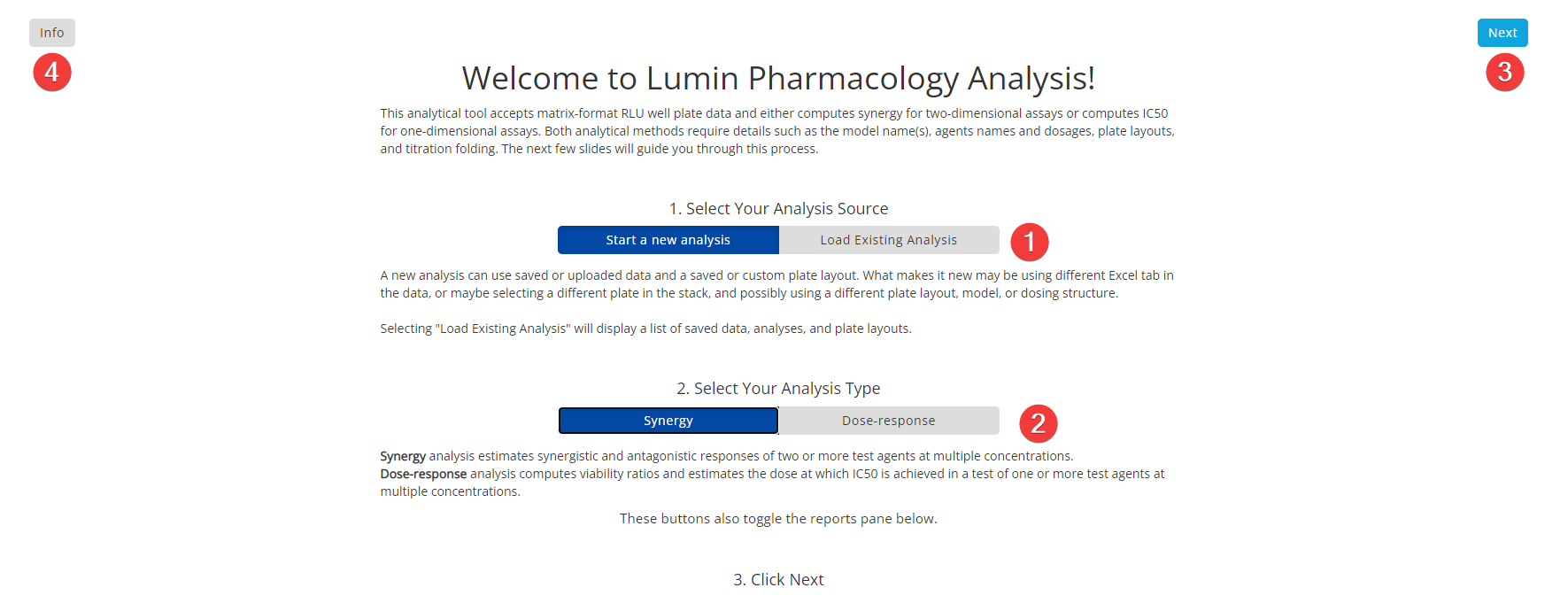
Step 1: Introduction and Analysis Type
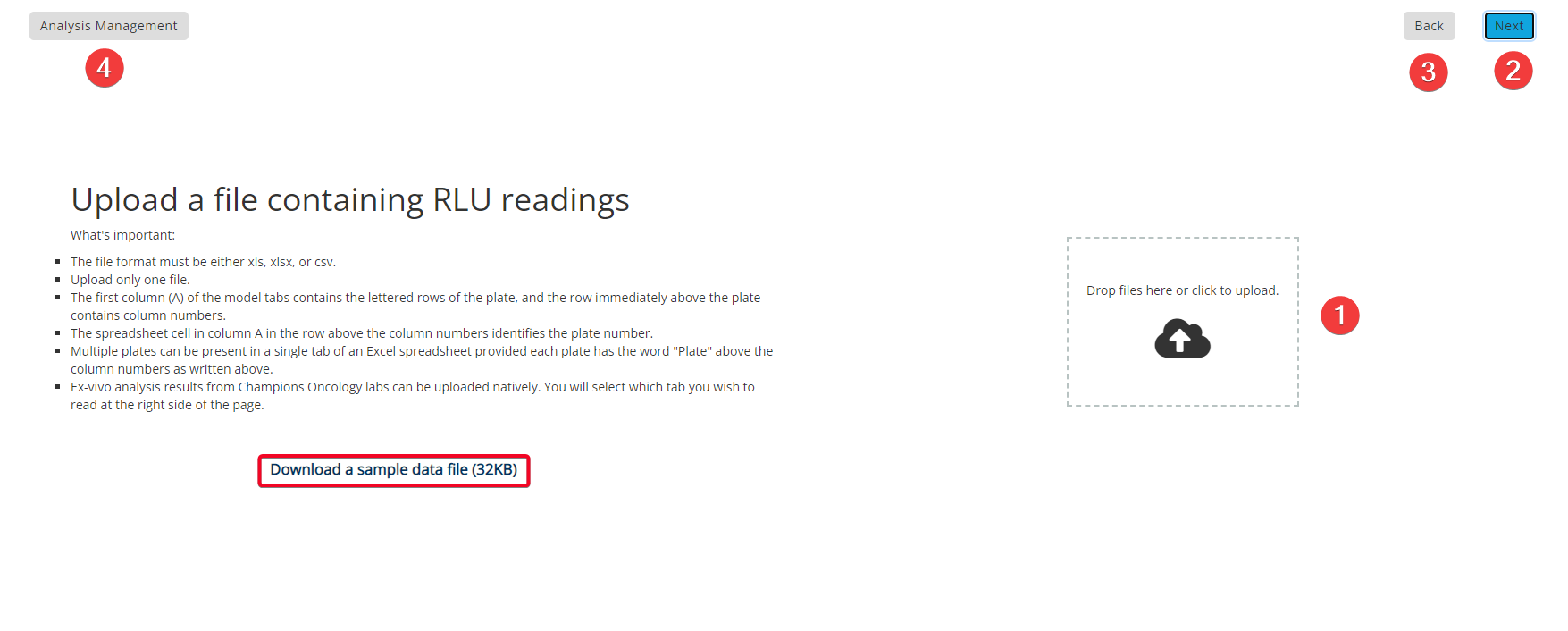
Step 2: Upload File
You can also download a sample file by clicking the text that is highlighted in the red box above.
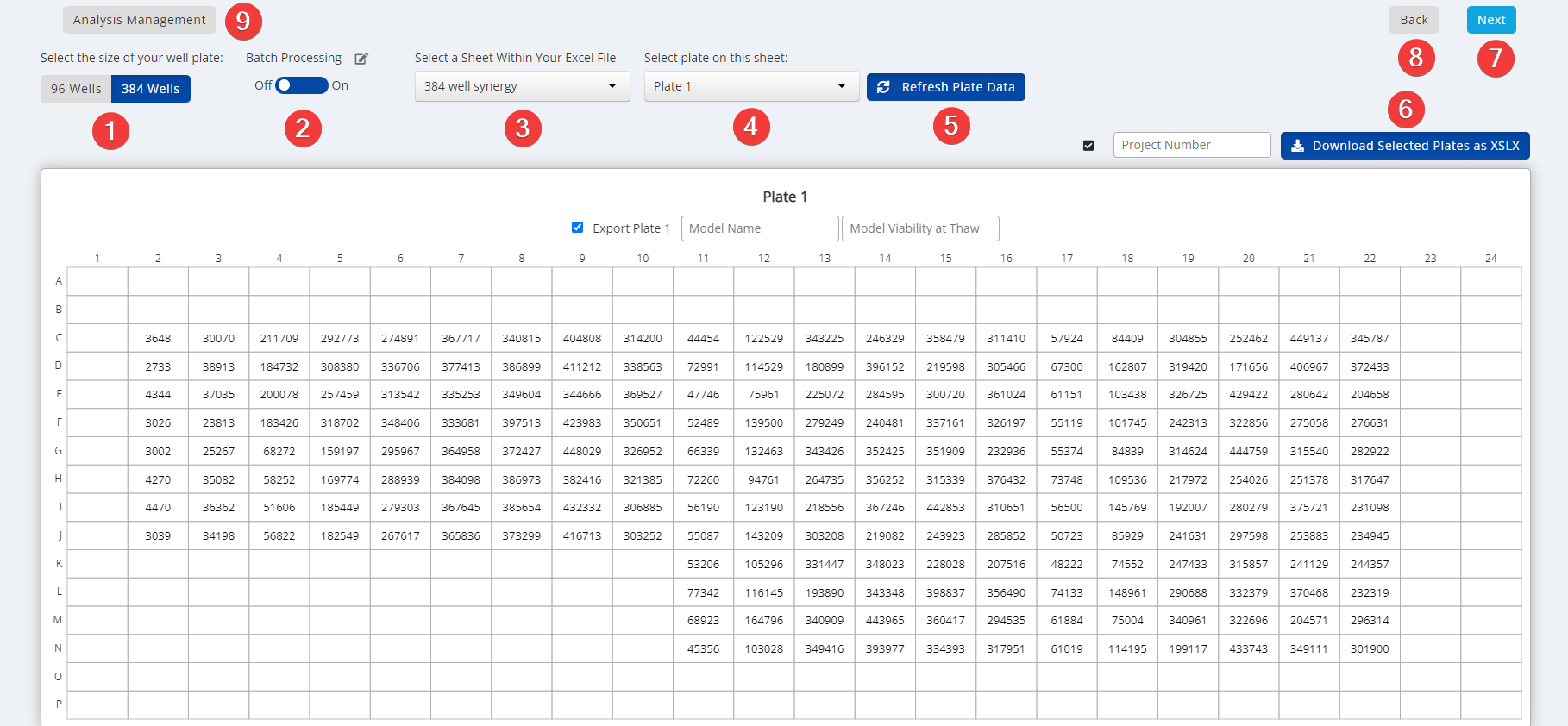
Step 3: Select Plate
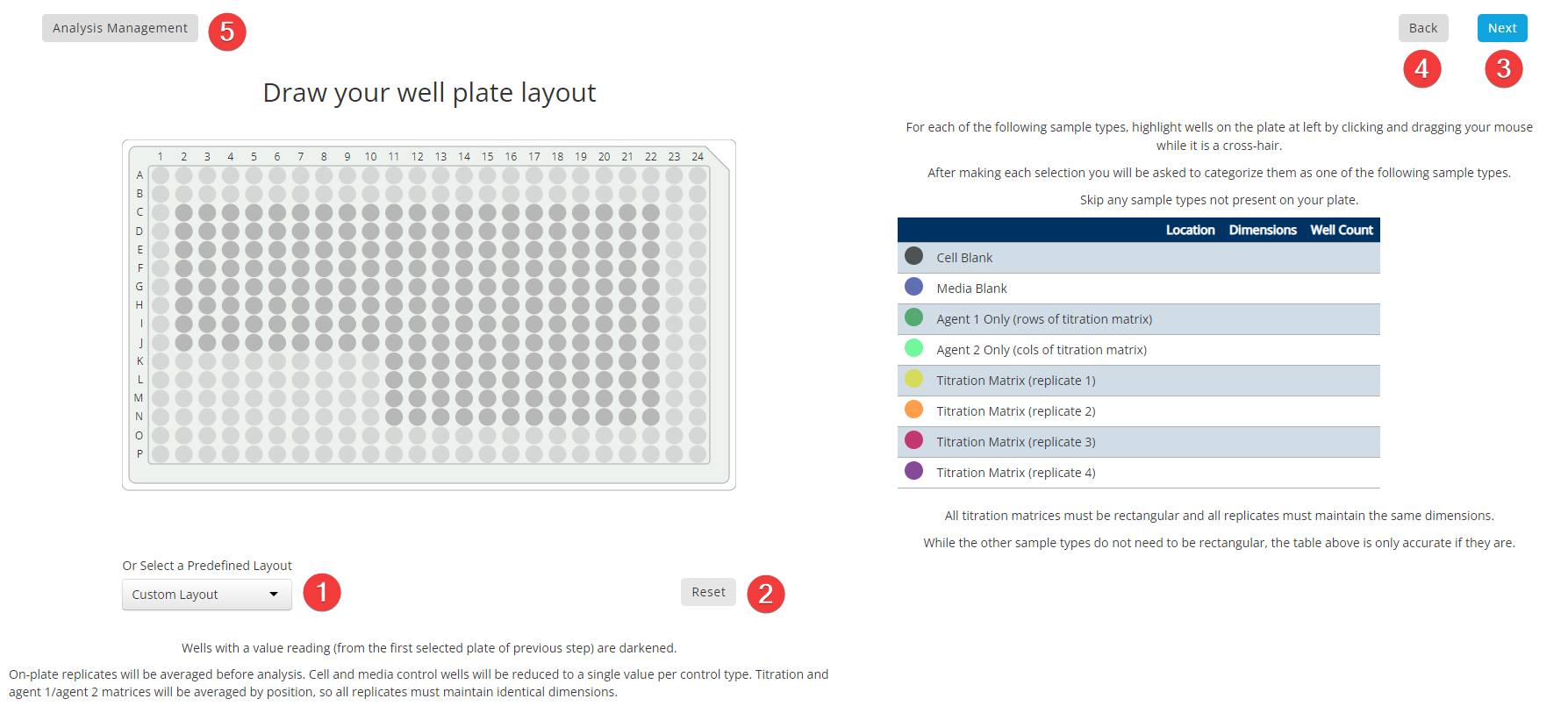
Step 4: Plate Layout

Synergy Step 5: Plate Titration
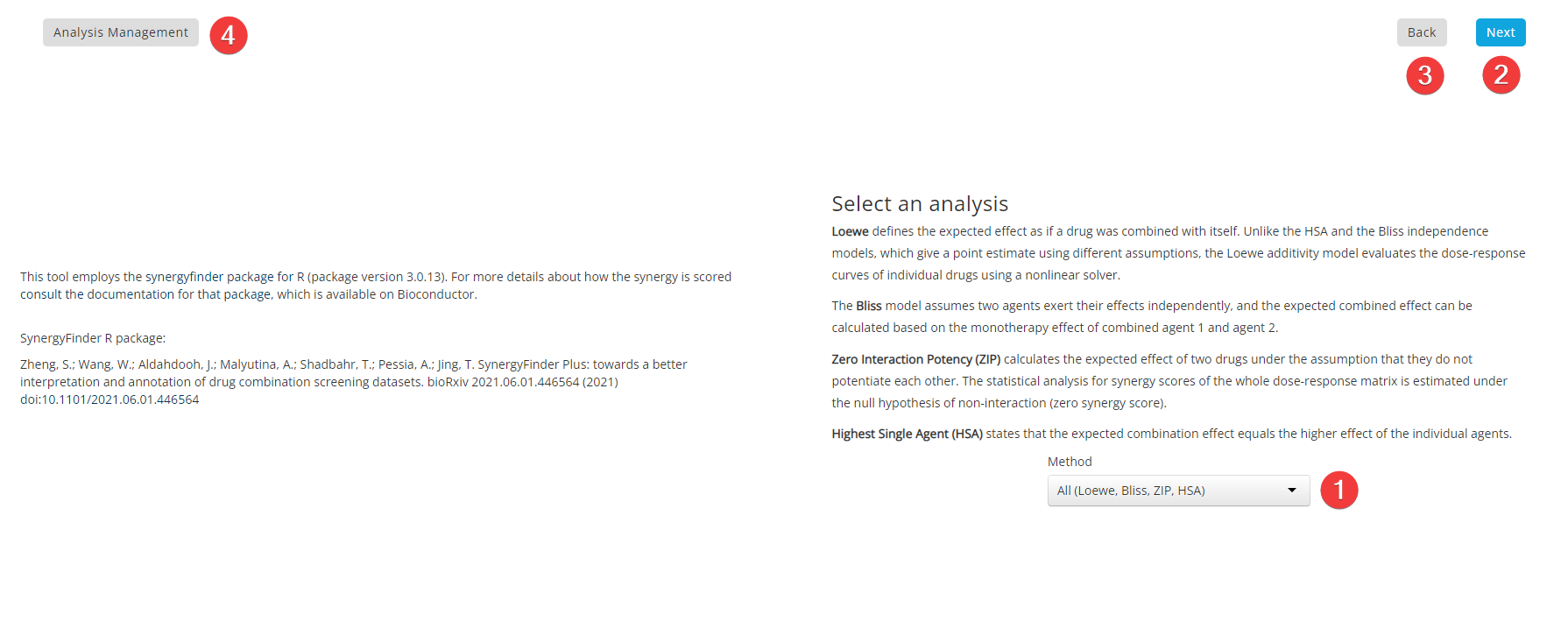
Synergy Step 6: Analysis Parameters
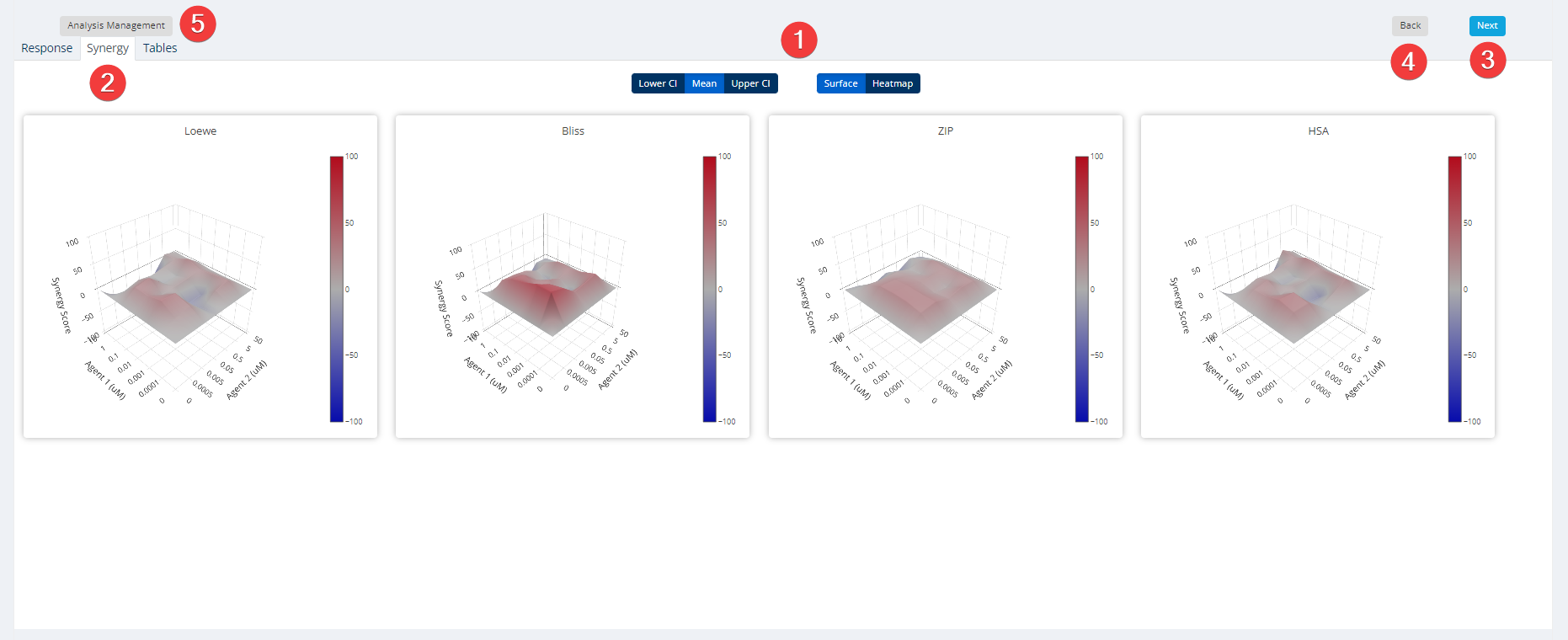
Synergy Step 7: Results & Report
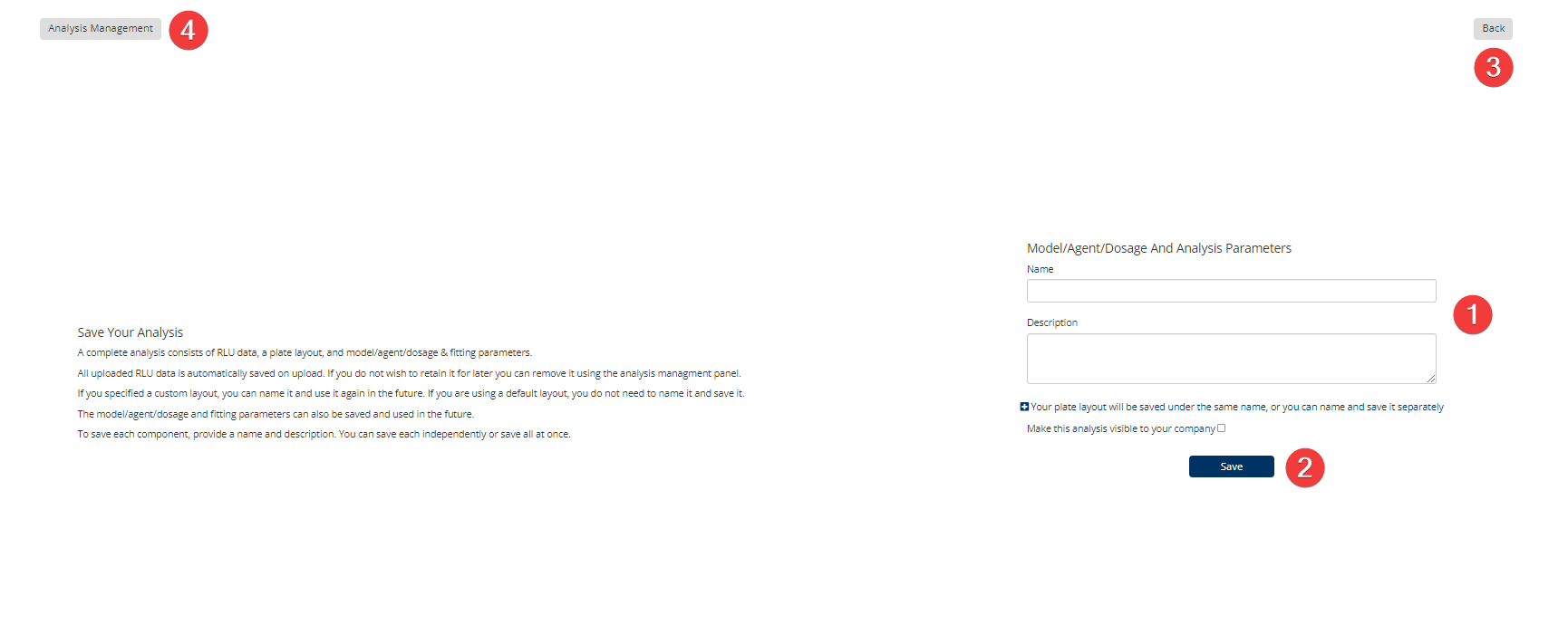
Synergy Step 8: Save
The summary synergy scores can be interpreted as the average excess response due to drug interactions (i.e. synergy score of 15 corresponds to 15% of response beyond expectation). There is no particular threshold to define a good synergy score, since combination synergy is highly context-specific. However, based on our experience, the synergy scores near 0 gives limited confidence on antagonism or synergy. So when a synergy score is:
Similar thresholds can be applied to the most synergistic area (MSA) score, which represents the most synergistic 3-by-3 dose-window in a dose-response matrix.

Dose Response Step 5: Plate Titration

Dose Response Step 6: Analysis Parameters
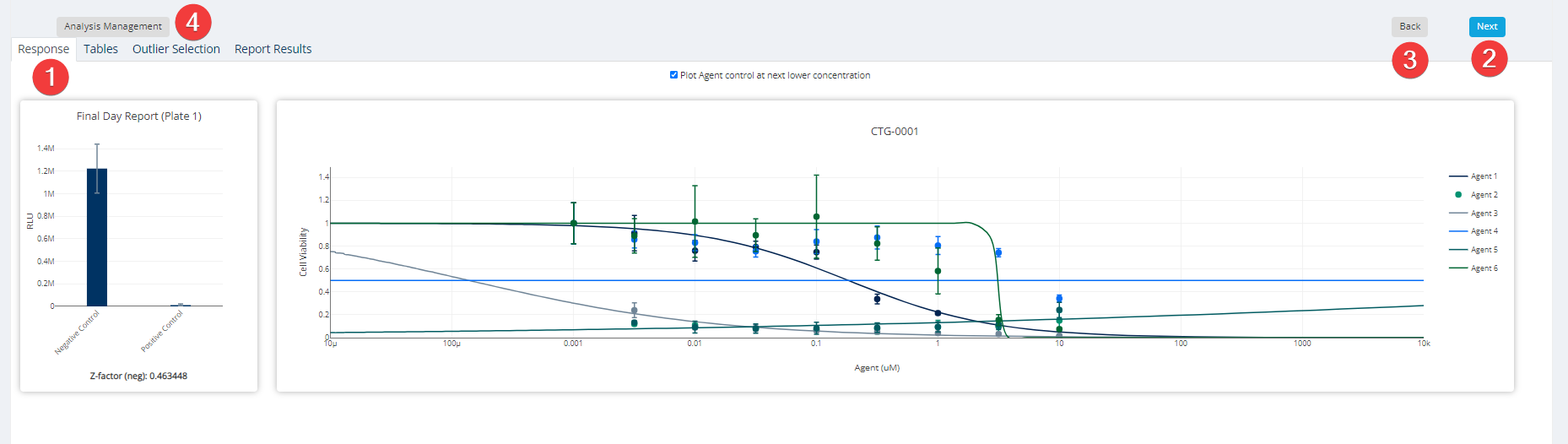
Dose Response Step 7: Results & Report
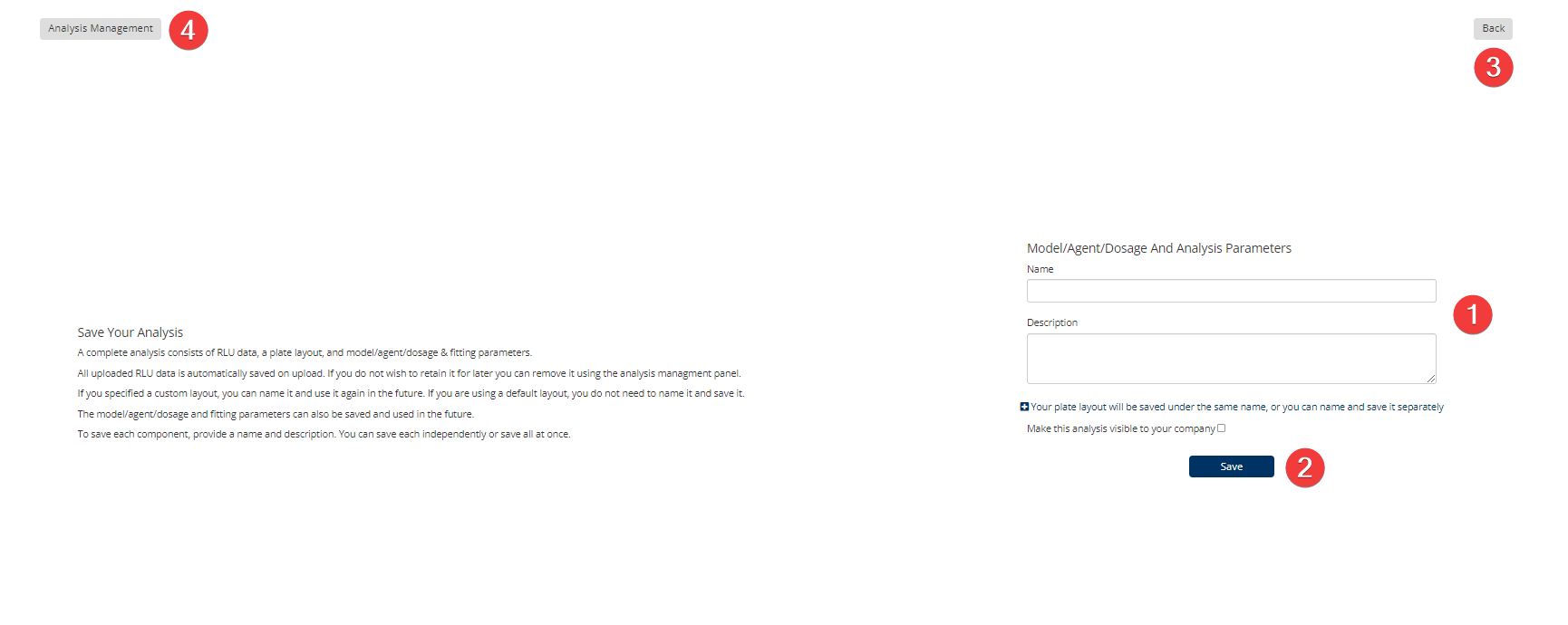
Dose Response Step 8: Save
Many people who use the Lumin dose response calculator have performed similar analyses in Graphpad Prism and are curious to know how our IC50 values compare to those computed in Prism, or they have actually done a comparison and are attempting to explain the difference between the two sets of results. During development and testing we have done our best to “tune” Lumin to produce similar results as Prism where possible. This is possible because both Prism and the Lumin Pharmacology analysis tool fit a fairly well-documented 4-parameter log-logistic model (LL.4).
Prior to fitting the LL.4 Lumin removes the background luminance by subtracting the mean media control reading from each observation. It then computes the viability ratio relative to the control as observed/vehicle control. The viability ratios are then moved to model fitting. The model fitting in Lumin is done by the software program R using the drc package (see reference below). The 4 parameters in the model are the bottom, the top, the slope, and the IC50. In Lumin the default is to fix the bottom at 0 and the top at 1, although it is possible to unset these and have the model fit them in addition to the IC50 and the slope. Lumin also uses a mean-least-squares estimation method and the default starting functions for the optimization method, but it is possible to override the start values for both IC50 and slope if necessary. Beyond the option to unfix the bottom and/or top and set start values, the only other deviation from the drc defaults is to set the optimization method to “Nelder-Mead” rather than “BFGS”, as we have observed this method produces output that match Prism to 4 significant figures. Lumin also has a configurable default of 1000 maximum iterations. When we observe a difference between Lumin and Prism it is usually due to higher than ideal variance in the data. In these cases, the stochastic nature of the optimization algorithm leaves the door open for two optimization methods to converge at slightly different IC50 estimates within acceptable relative tolerance, due either to using different relative tolerance values or to using different hop sizes that may permit them to identify different local minimum fit estimators rather than a single global minimum. The default value for relative convergence in drc is 1e-7, and the dose-scaling and response-scaling thresholds are both set to 1e-15. The following table lists the default optimization and LL.4 model settings for Lumin and Prism.
| Parameter | Lumin | Prism |
|---|---|---|
| Bottom | 0* | 0* |
| Top | 1* | 1* |
| IC50 start | automatic* | automatic* |
| Slope start | automatic* | automatic* |
| Regression method non-robust | mean least squares | non-robust least squares* |
| Convergence criteria | 1e-7 | Medium/strict* |
| Maximum iterations | 1000* | 1000* |
| Outlier test | Grubbs | Grubbs/ROUT* |
| Optimization method | Nelder-Mead | Marquardt |
*User configurable
Two and three-agent synergy calculations are done in R using the package synergyfinder. This package is exceptionally well documented by the authors (see references below), and Lumin users should refer to that documentation for descriptions of the methods. Prior to passing the observed values to synergyfinder, Lumin pre-processes the data by subtracting the mean background reading (the media control wells) from each observed value, then it computes the percent inhibition as 1 - observed/vehicle control. All synergy estimations are based on the inhibition metric.
Select the ‘Pharmacology Analysis’ Tool from the Lumin Homepage.
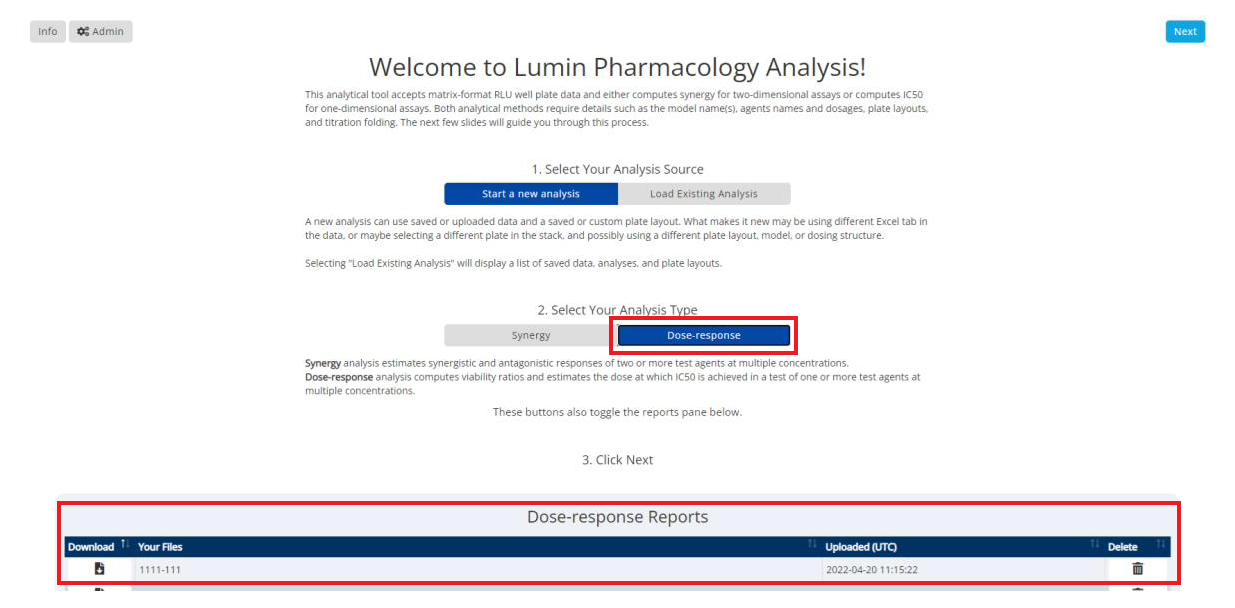
Under “2. Select Your Analysis Type”, choose Dose Response. Your report(s) will be listed underneath Dose-response Reports at the bottom of the screen, click the download button to export the PDF.
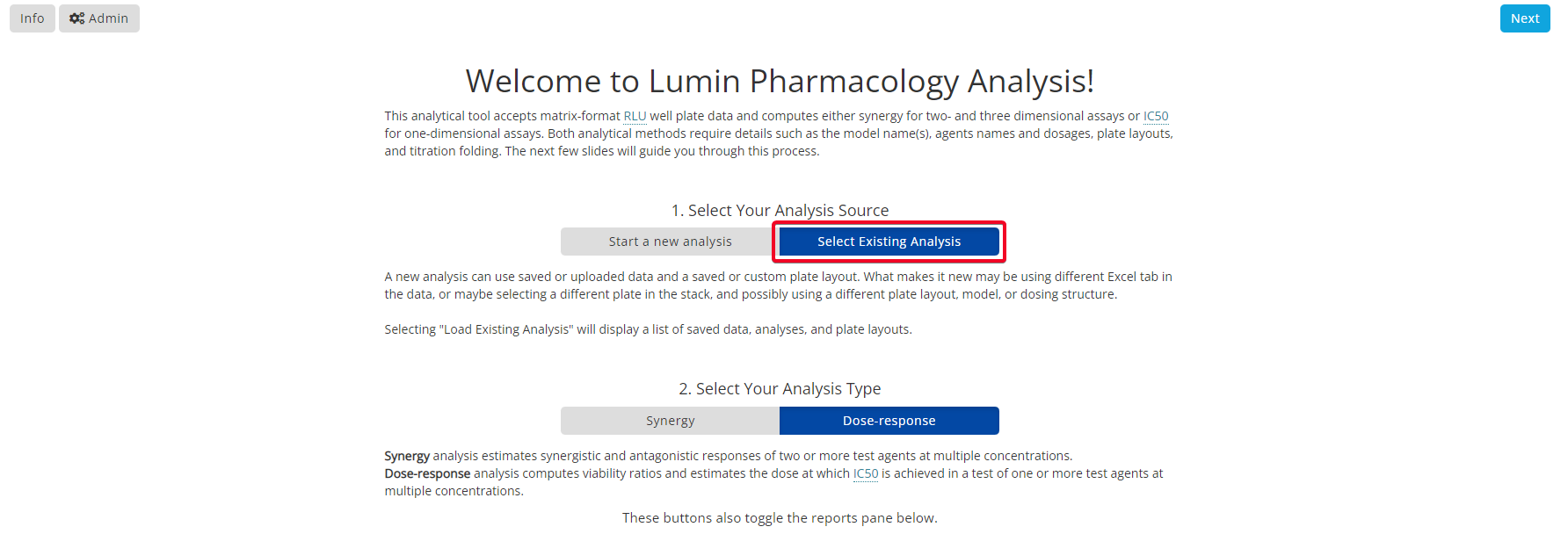
To download the raw RLU values, on the same page, select ‘Load Existing Analysis’. Then select ‘Data Files’ on the left hand side menu. Next, select the data file(s) to download.
Loewe defines the expected effect as if a drug was combined with itself. Unlike the HSA and the Bliss independence models, which give a point estimate using different assumptions, the Loewe additivity model evaluates the dose-response curves of individual drugs using a nonlinear solver.
The Bliss model assumes two agents exert their effects independently, and the expected combined effect can be calculated based on the monotherapy effect of combined agent 1 and agent 2.
Zero Interaction Potency (ZIP) calculates the expected effect of two drugs under the assumption that they do not potentiate each other. The statistical analysis for synergy scores of the whole dose-response matrix is estimated under the null hypothesis of non-interaction (zero synergy score).
Highest Single Agent (HSA) states that the expected combination effect equals the higher effect of the individual agents.
Grubbs’s Test - This is a test used to detect outliers in a univariate data set assumed to come from a normally distributed population (also known as the Maximum Normed Residual Test).
Synergy Analysis is based on the work of Zheng et al: Zheng, S.; Wang, W.; Aldahdooh, J.; Malyutina, A.; Shadbahr, T.; Pessia, A.; Jing, T. SynergyFinder Plus: towards a better interpretation and annotation of drug combination screening datasets. bioRxiv 2021.06.01.446564 (2021) doi:10.1101/2021.06.01.446564
Synergy Analysis Interpretation is based on the (SynergyFinder - Documentation, 2022): Synergyfinder.fimm.fi. 2022. SynergyFinder - Documentation. [online] Available at: https://synergyfinder.fimm.fi/synergy/synfin_docs/ [Accessed 2022].
Dose-Response Analysis is based on the work of Ritz et al: Ritz C, Baty F, Streibig JC, Gerhard D (2015) Dose-Response Analysis Using R. PLoS ONE 10(12): e0146021. doi:10.1371/journal.pone.0146021 or http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0146021